
What it is not

A normalized, well-structured, relational database that you could query like:
Give me all pH - values where we also have turbidity measurements and discharge is < 20 m3/s.
And plot them. In blue. With yellow dots. On a logarithmic scale.
- That would be impossible, EAWAG-wide.
- That doesn't solve the problem (archival).
What is it for?
A motivating example:
On Tue, Aug 25, 2015 at 6:27 PM, Matthew MacLeod <Matthew.MacLeod@aces.su.se>
wrote:
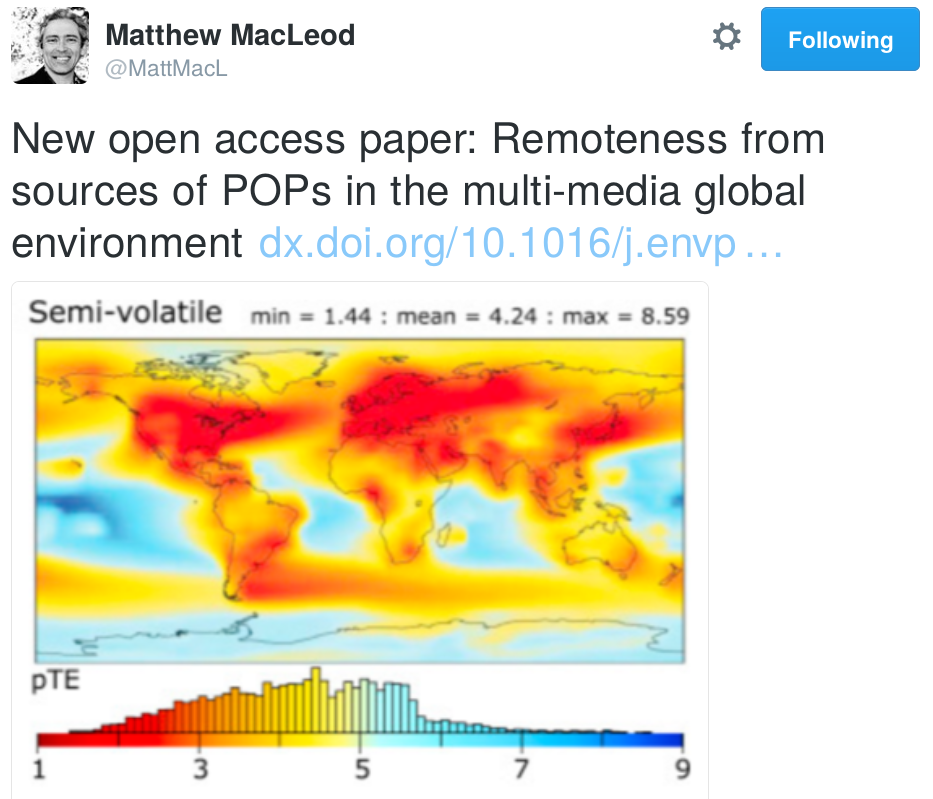
Hey HvW…
Recep and I have been working a bit on some ideas related to your
Remoteness Index.
It would help us a lot to have the raw data (as NetCDF files, for
example) that underlie your Remoteness Index maps that you
published in ES&T. We are most interested in the map for the
night-light emission scenario, but would be happy to have the raw
data for the CROP scenario also.
Do you still have those files backed up somewhere??
Thanks! -matt

Data Mangement Rule at the time:
- Put your stuff on a CD.
- Put CD into drawer in secretary's office.
- CD not extremely good organized and documented.
- Who in old workgroup would be willing to search and interpret that CD? (In case it it still working)
- Workgroup will cease to exist soon.
My private data management strategy at the time:
rsync -av /home/hvwaldow hvw@climstor.unibe.ch:/.../private/archive/phd
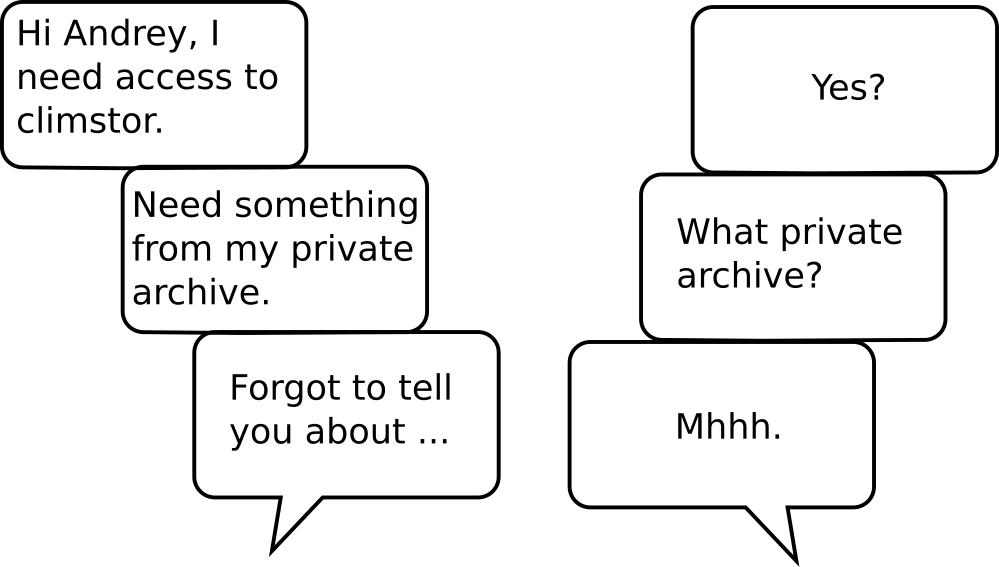
recovery ...
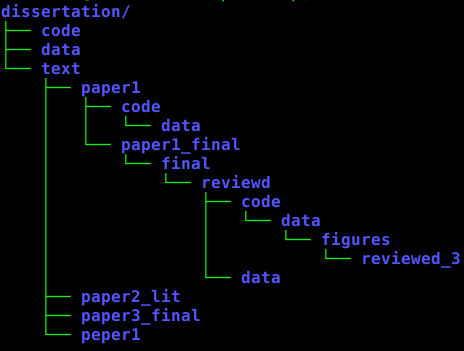
recovery ...
- find file (candidates).
- don't find plotting routine.
- re-write plotting routine.
- compare with published figure.
- document data
On Wed, Sep 16, 2015 at 2:30 AM, Harald von Waldow <harald@vonwaldow.ch> wrote:
Hi Matt & Recep,
sorry for the delay again. You know how it is ..
Please find attached an archive with 4 files:
.....
....Feb 1, 2016:
Happy End
What data to upload?
E.g.
- data that supplements a publication
- raw - data
- the workgroup "stock-data"
- ...
- ...
Rule of thumb: Everything that is consistent and complete enough and worth checking and documenting to an extent that it stands for itself and could be used by others without additional information.
Details are going to be figured out individually.
What is it then, exactly?
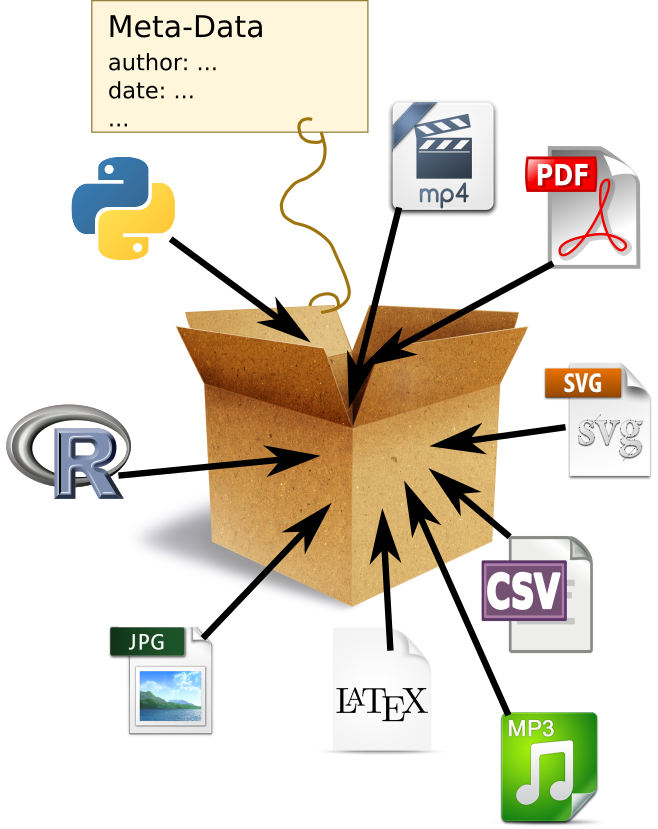
A collection
of
"Data
Packages"
The system doesn't look into the files.
no selection/search/analysis based directly on file content.
technically, everything can be dumped into a package, without any pre-processing.

 Some file-formats should be avoided, though!
Some file-formats should be avoided, though!
date of submission: ...
temporal coverage: ...
spatial coverage: ...
status: ...
Variables: ...
Systems: ...

package = files + meta-data
"resources" = files
- search for packages is based on meta-data.
- interaction with other repositories and services relies on meta-data.
- other "meta-data", e.g., units of variables, should go into a resource (= file).
How is it organized?
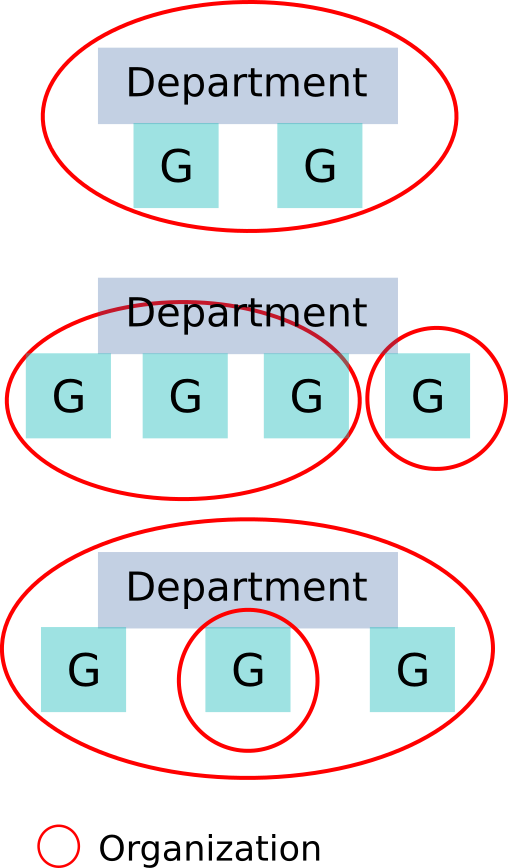
=
Departments
or
Research Groups
Organizations are, ideally, comparatively homogeneous with respect to data-management needs.
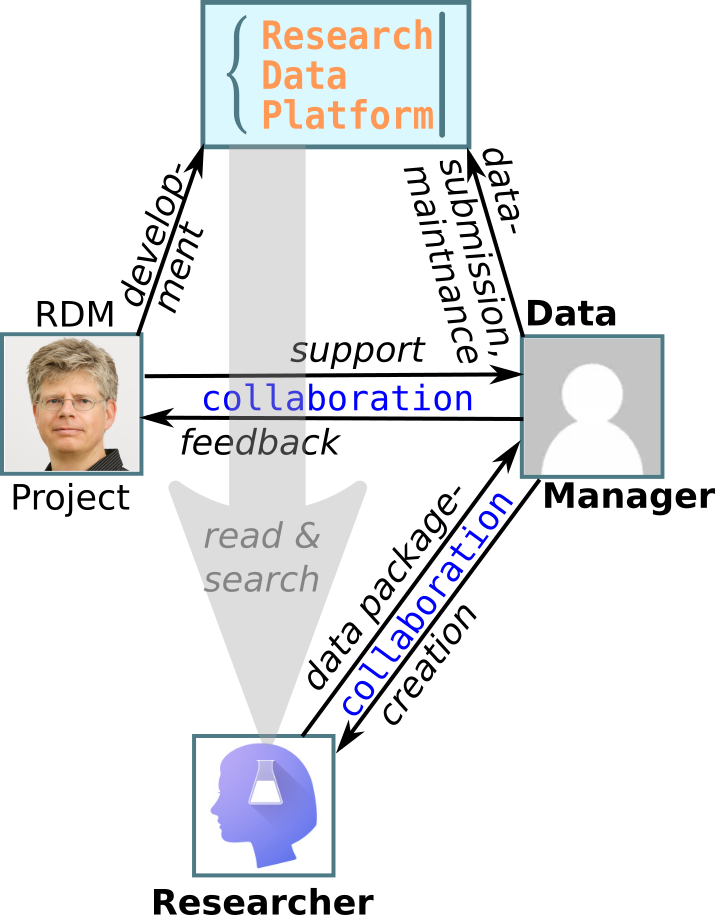
"Data Managers"
- are the link between researchers, the platform, and the project.
- are RDP - administrators for their organization.
- number preferably 1 - 3 per organization.
- ideally have
- some data-handling skills.
- prolonged stay at Eawag.
- good overview of organizations' research-activities.
How does it look like?
What about automating things?
CKAN has a remote API that exposes all of CKAN’s core features.
Access via http:
http://eaw-ckan-dev1/api/3/action/package_search?fq=tags:fish
Used to programmatically read, create and modify packages.
- recuring tasks, e.g. regular updates
- batch upload of legacy data
- automatic validation
- interaction with other repositories
# sh
curl 'http://eaw-ckan-dev1.eawag.wroot.emp-eaw.ch/api/3/action/organization_list'
{"help": ... "result": ["aquatic-ecology", "aquatic-entomology",
"environmental-chemistry", "environmental-microbiology",
"fish-ecology-and-evolution", "gis-services", "it-services", ... ] }# Python
from ckanapi import *
ckanremote = RemoteCKAN("http://eaw-ckan-dev1.eawag.wroot.emp-eaw.ch",
apikey=os.environ["CKAN_DEV1_APIKEY_HVW_ADM"])
pkg = ckanremote.action.package_show(id="reform")
print(pkg)## R
> library('ckanr')
> ckanr_setup(url="http://eaw-ckan-dev1",key=Sys.getenv("CKAN_DEV1_APIKEY_HVW_ADM"))
> package_list(as="table")
[1] "data-from-population..."
[2] "record"
...
Support for writing such client code expected as major task during pilot-phase.
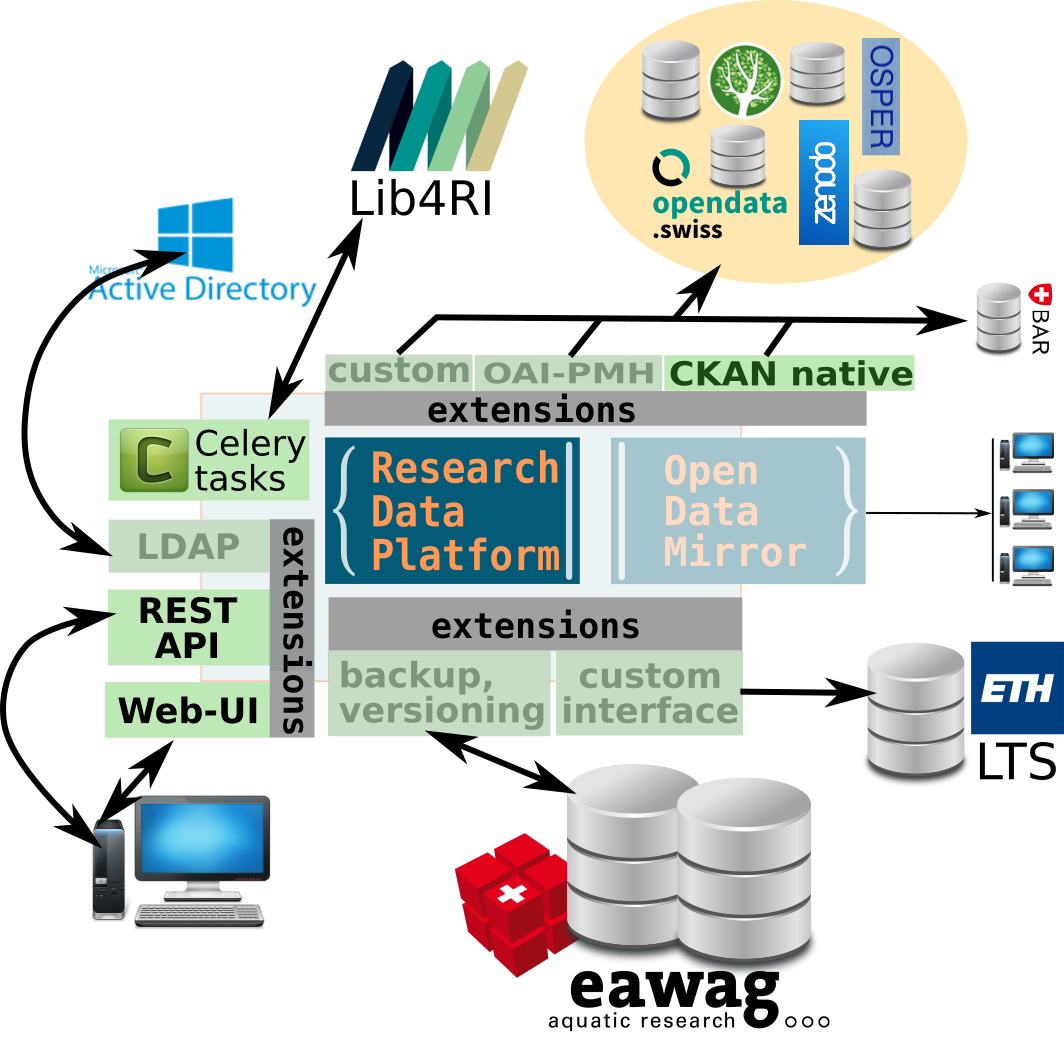
How does it fit into the technical ecosystem?
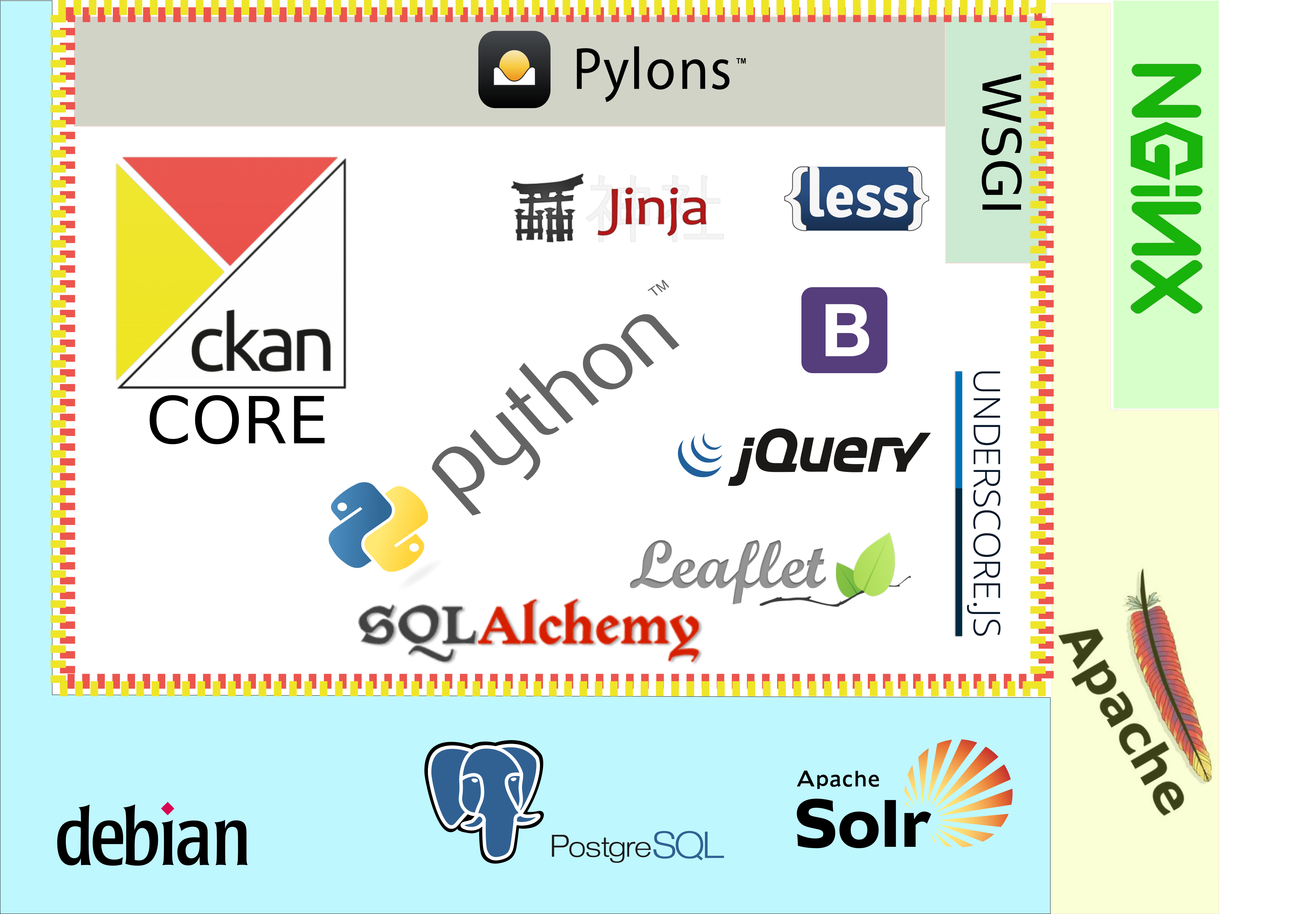
Whatsoftware is that based on?
Time-line & Outlook
March 21 — Dec 31: Pilot-Phase
- Experimentation with various types of data.
- Debugging.
- User feedback for further iterative RDP development.
- User-specific development work, e.g., client-side helper tools.
- Collaborative development of workflows.
- No data-safety!
- No interface-stability!
New features until 2017
- Significant development is planned to take place during this year.
- Some
features are already planned, but concrete
implementation and yet unknown needs will
emerge. E.g.:
- version control for resources
- strong backup
- interfacing with public repositories (Zenodo, Dryad, ...)
- public facing server / Open Data
- ldap authentification
- user-friendly spatial input
- ...
Next steps
- Data-Managers, email me: harald.vonwaldow@eawag.ch
I would like to be able to associate each group with a name by May 15, 2016. - We will have workshops where I coach you through data-submission and collect your requirements.
- Everybody: Search and download, submit bugs and requests:
https://github.com/eawag-rdm/eawag-ckan/issues. - Have your say in the development: Subscribe
to
eaw-rdm-discuss@sympa.ethz.ch
Questions?
